class: center, middle, inverse, title-slide .title[ # Module 1: Hospital Pricing and Selection on Observables ] .subtitle[ ## Part 2: Matching and Weighting ] .author[ ### Ian McCarthy | Emory University ] .date[ ### Econ 470 & HLTH 470 ] --- <!-- Adjust some CSS code for font size and maintain R code font size --> <style type="text/css"> .remark-slide-content { font-size: 30px; padding: 1em 2em 1em 2em; } .remark-code { font-size: 15px; } .remark-inline-code { font-size: 20px; } </style> <!-- Set R options for how code chunks are displayed and load packages --> # Goal Find covariates `\(X_{i}\)` such that the following assumptions are plausible: 1. Selection on observables: `$$Y_{0i}, Y_{1i} \perp\!\!\!\perp D_{i} | X_{i}$$` 2. Common support: `$$0 < \text{Pr}(D_{i}=1|X_{i}) < 1$$` -- Then we can use `\(X_{i}\)` to group observations and use expectations for control as the predicted counterfactuals among treated, and vice versa. --- # Assumption 1: Selection on Observables `\(E[Y_{1}|D,X]=E[Y_{1}|X]\)` -- In words...nothing unobserved that determines treatment selection and affects your outcome of interest. --- # Assumption 1: Selection on Observables - Example of selection on observables from *Mastering Metrics* --- # Assumption 2: Common Support Someone of each type must be in both the treated and untreated groups -- `$$0 < \text{Pr}(D=1|X) <1$$` --- # Causal inference with observational data With selection on observables and common support: 1. Subclassification 2. Matching estimators 3. Reweighting estimators 4. Regression estimators --- # Subclassification Sum the average treatment effects by group, and take a weighted average over those groups: `$$ATE=\sum_{i=1}^{N} P(X=x_{i}) \left(E[Y | X, D=1] - E[Y | X, D=0]\right)$$` --- # Subclassification - Difference between treated and controls - Weighted average by probability of given group (proportion of sample) - What if outcome is unobserved for treatment or control group for a given subclass? -- - This is the *curse of dimensionality* --- # Matching: The process 1. For each observation `\(i\)`, find the `\(m\)` "nearest" neighbors, `\(J_{m}(i)\)`. 2. Impute `\(\hat{Y}_{0i}\)` and `\(\hat{Y}_{1i}\)` for each observation:<br> `$$\hat{Y}_{0i} = \begin{cases} Y_{i} & \text{if} & D_{i}=0 \\ \frac{1}{m} \sum_{j \in J_{m}(i)} Y_{j} & \text{if} & D_{i}=1 \end{cases}$$` `$$\hat{Y}_{1i} = \begin{cases} Y_{i} & \text{if} & D_{i}=1 \\ \frac{1}{m} \sum_{j \in J_{m}(i)} Y_{j} & \text{if} & D_{i}=0 \end{cases}$$` 3. Form "matched" ATE:<br> `\(\hat{\delta}^{\text{match}} = \frac{1}{N} \sum_{i=1}^{N} \left(\hat{Y}_{1i} - \hat{Y}_{0i} \right)\)` --- # Matching: Defining "nearest" 1. Euclidean distance:<br> `\(\sum_{k=1}^{K} (X_{ik} - X_{jk})^{2}\)` 2. Scaled Euclidean distance:<br> `\(\sum_{k=1}^{K} \frac{1}{\sigma_{X_{k}}^{2}} (X_{ik} - X_{jk})^{2}\)` 3. Mahalanobis distance:<br> `\((X_{i} - X_{j})' \Sigma_{X}^{-1} (X_{i} - X_{j})\)` --- # Animation for matching .center[  ] --- # Matching: Defining "nearest" - But are observations really the same in each group? - Potential for "matching discrepancies" to introduce bias in estimates -- - "Bias correction" based on `$$\hat{\mu}(x_{i}) - \hat{\mu}(x_{j(i)})$$` (i.e., difference in fitted values from regression of `\(y\)` on `\(x\)`, with the difference between observed `\(Y_{1i}\)` and imputed `\(Y_{0i}\)`) --- # Weighting 1. Estimate propensity score `ps <- glm(D~X, family=binomial, data)`, denoted `\(\hat{\pi}(X_{i})\)` 2. Weight by inverse of propensity score<br> .center[ `\(\hat{\mu}_{1} = \frac{ \sum_{i=1}^{N} \frac{Y_{i} D_{i}}{\hat{\pi}(X_{i})} }{ \sum_{i=1}^{N} \frac{D_{i}}{\hat{\pi}(X_{i})} }\)` and `\(\hat{\mu}_{0} = \frac{ \sum_{i=1}^{N} \frac{Y_{i} (1-D_{i})}{1-\hat{\pi}(X_{i})} }{ \sum_{i=1}^{N} \frac{1-D_{i}}{1-\hat{\pi}(X_{i})} }\)` ] 3. Form "inverse-propensity weighted" ATE:<br> .center[ `\(\hat{\delta}^{IPW} = \hat{\mu}_{1} - \hat{\mu}_{0}\)` ] --- # Regression 1. Regress `\(Y_{i}\)` on `\(X_{i}\)` among `\(D_{i}=1\)` to form `\(\hat{\mu}_{1}(X_{i})\)` 2. Regress `\(Y_{i}\)` on `\(X_{i}\)` among `\(D_{i}=0\)` to form `\(\hat{\mu}_{0}(X_{i})\)` 3. Form difference in predictions:<br> .center[ `$$\hat{\delta}^{reg} = \frac{1}{N} \sum_{i=1}^{N} \left(\hat{\mu}_{1}(X_{i}) - \hat{\mu}_{0}(X_{i})\right)$$` ] --- # Regression Or estimate in one step, .center[ `$$Y_{i} = \delta D_{i} + \beta X_{i} + D_{i} \times \left(X_{i} - \bar{X}\right) \gamma + \varepsilon_{i}$$` ] -- - Note the `\((X_{i} - \bar{X})\)`. What does this do? --- # Animation for regression .center[ 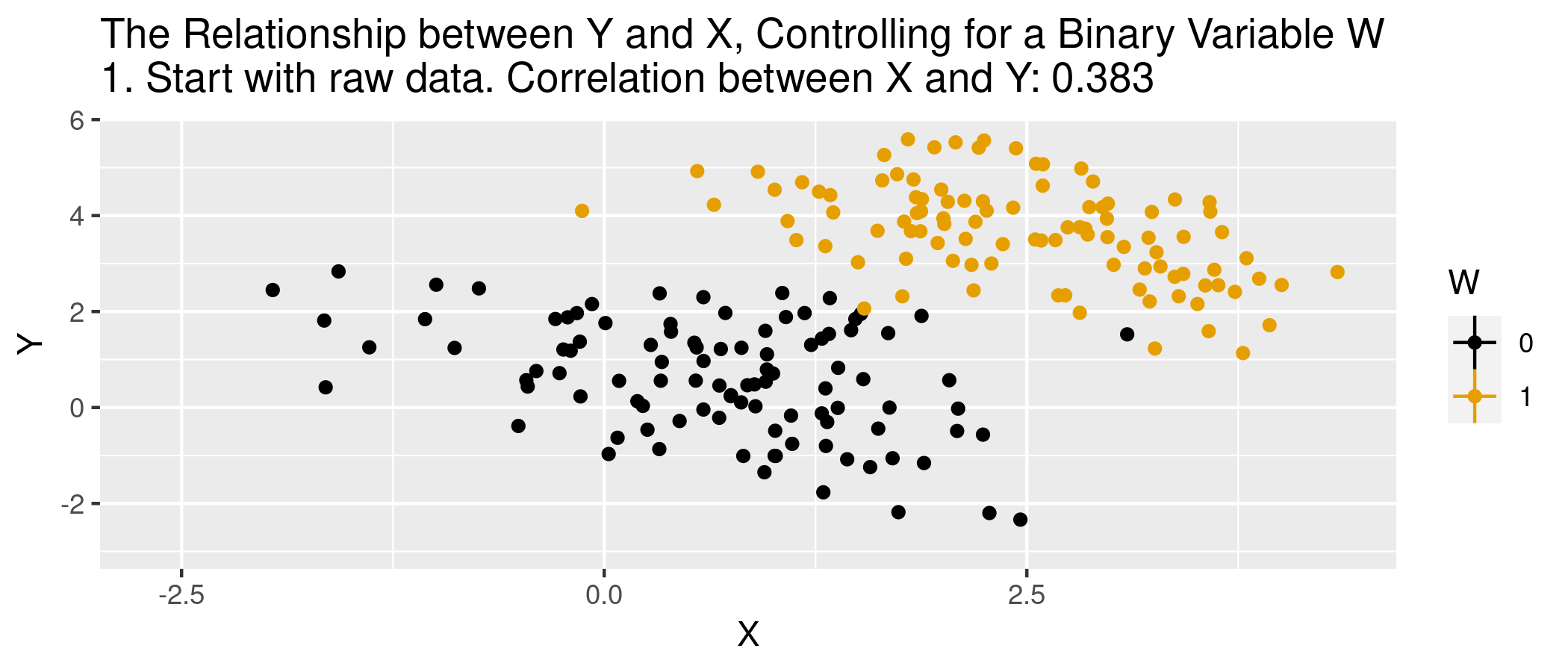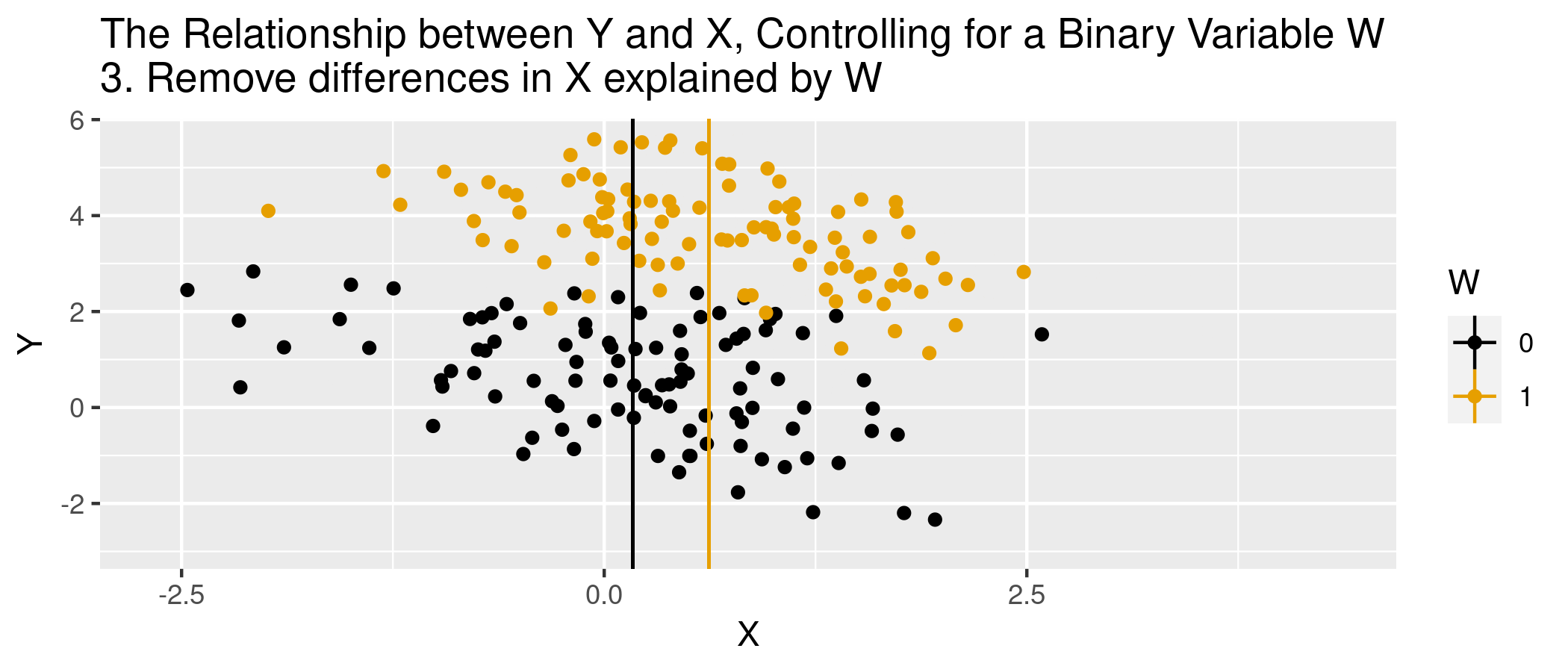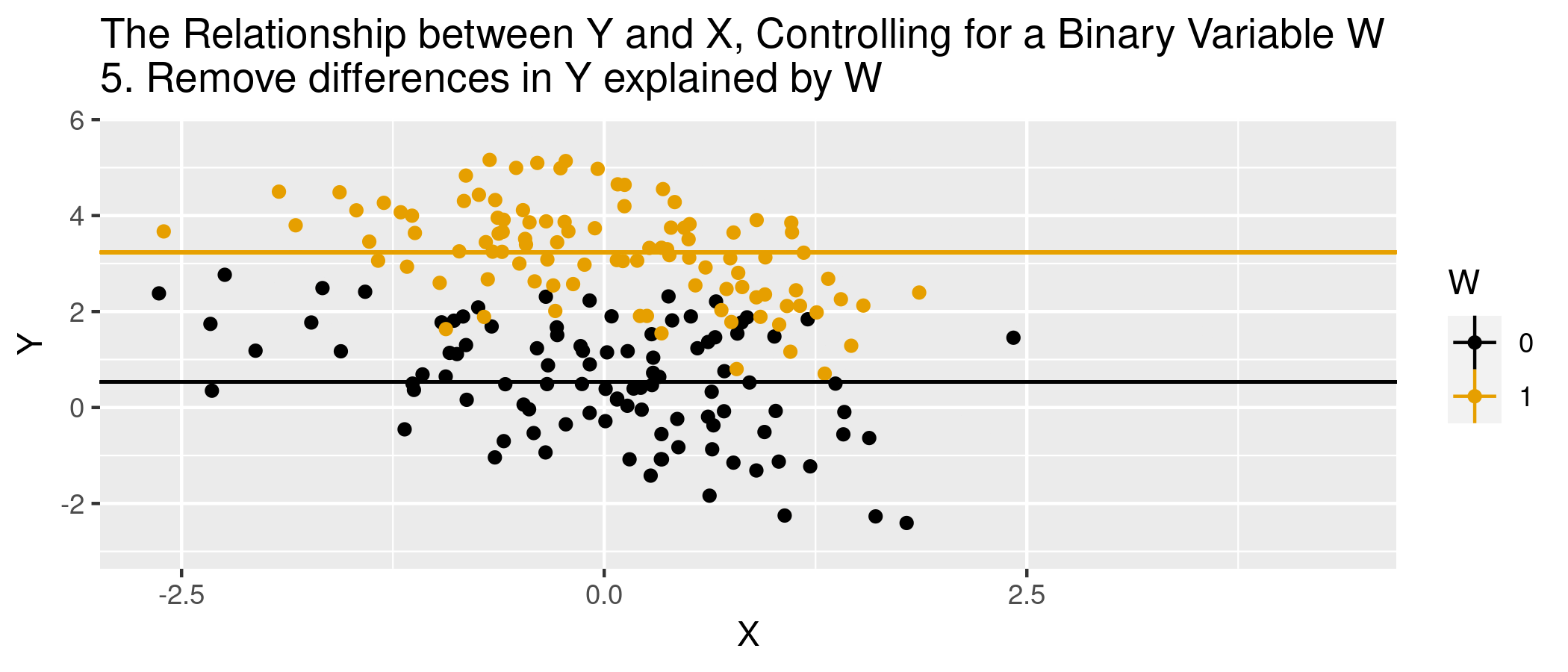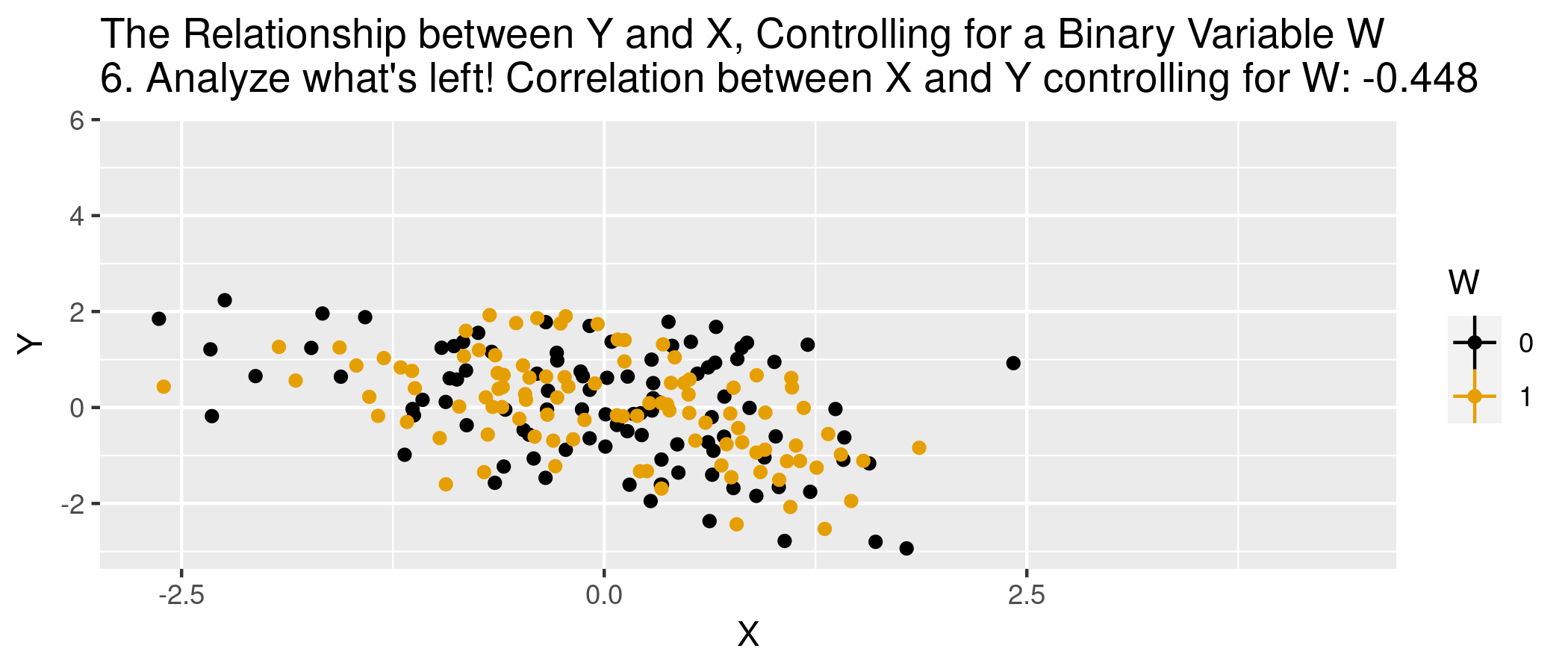 ] --- # Simulated data Now let's do some matching, re-weighting, and regression with simulated data: ```r n <- 5000 select.dat <- tibble( x = runif(n, 0, 1), z = rnorm(n, 0, 1), w = (x>0.65), y = -2.5 + 4*w + 1.5*x + rnorm(n,0,1), w_alt = ( x + z > 0.35), y_alt = -2.5 + 4*w_alt + 1.5*x + 2.25*z + rnorm(n,0,1) ) ``` --- # Simulation: nearest neighbor matching ```r nn.est1 <- Matching::Match(Y=select.dat$y, Tr=select.dat$w, X=select.dat$x, M=1, Weight=1, estimand="ATE") summary(nn.est1) ``` ``` ## ## Estimate... 3.8785 ## AI SE...... 0.53145 ## T-stat..... 7.298 ## p.val...... 2.9199e-13 ## ## Original number of observations.............. 5000 ## Original number of treated obs............... 1731 ## Matched number of observations............... 5000 ## Matched number of observations (unweighted). 5013 ``` --- # Simulation: nearest neighbor matching ```r nn.est2 <- Matching::Match(Y=select.dat$y, Tr=select.dat$w, X=select.dat$x, M=1, * Weight=2, estimand="ATE") summary(nn.est2) ``` ``` ## ## Estimate... 3.8785 ## AI SE...... 0.53145 ## T-stat..... 7.298 ## p.val...... 2.9199e-13 ## ## Original number of observations.............. 5000 ## Original number of treated obs............... 1731 ## Matched number of observations............... 5000 ## Matched number of observations (unweighted). 5013 ``` --- # Simulation: regression ```r reg1.dat <- select.dat %>% filter(w==1) reg1 <- lm(y ~ x, data=reg1.dat) reg0.dat <- select.dat %>% filter(w==0) reg0 <- lm(y ~ x, data=reg0.dat) pred1 <- predict(reg1,new=select.dat) pred0 <- predict(reg0,new=select.dat) mean(pred1-pred0) ``` ``` ## [1] 4.126236 ``` --- # Violation of selection on observables .pull-left[ <u>NN Matching</u> ```r nn.est3 <- Matching::Match(Y=select.dat$y_alt, Tr=select.dat$w_alt, X=select.dat$x, M=1, Weight=2, estimand="ATE") summary(nn.est3) ``` ``` ## ## Estimate... 7.6502 ## AI SE...... 0.053248 ## T-stat..... 143.67 ## p.val...... < 2.22e-16 ## ## Original number of observations.............. 5000 ## Original number of treated obs............... 2756 ## Matched number of observations............... 5000 ## Matched number of observations (unweighted). 22555 ``` ] .pull-right[ <u>Regression</u> ```r reg1.dat <- select.dat %>% filter(w_alt==1) reg1 <- lm(y_alt ~ x, data=reg1.dat) reg0.dat <- select.dat %>% filter(w_alt==0) reg0 <- lm(y_alt ~ x, data=reg0.dat) pred1_alt <- predict(reg1,new=select.dat) pred0_alt <- predict(reg0,new=select.dat) mean(pred1_alt-pred0_alt) ``` ``` ## [1] 7.675315 ``` ] --- # What covariates to use? - There are such things as "bad controls" - We want to avoid control variables that are: -- - Outcomes of the treatment - Also endogenous (more generally)