ga_ma_2022 <- read_csv("../data/output/ma-snippets/ga-ma-data-2022.csv") %>%
select(-partc_score) %>% ungroup()
ma_ratings <- read_csv("../data/output/ma-snippets/ga-ratings-2022.csv")
ga_ma_full <- ga_ma_2022 %>%
left_join(ma_ratings, by="contractid") %>%
mutate(raw_rating=rowMeans(
cbind(breastcancer_screen, rectalcancer_screen, flu_vaccine,
physical_monitor, specialneeds_manage, older_medication, older_pain,
osteo_manage, diabetes_eye, diabetes_kidney, diabetes_bloodsugar,
ra_manage, falling, bladder, medication, statin, nodelays,
carequickly, customer_service, overallrating_care, overallrating_plan,
coordination, complaints_plan, leave_plan, improve, appeals_timely,
appeals_review, ttyt_available),
na.rm=T)) %>%
select(contractid, planid, fips, plan_type, partd, avg_enrollment, avg_eligibles,
avg_enrolled, premium, premium_partc, premium_partd, rebate_partc, ma_rate,
bid, avg_ffscost, partc_score, partcd_score, raw_rating)More Medicare Advantage Data
Ian McCarthy | Emory University
Outline for Today
- Revisit Medicare Advantage Data
- Examine Relationship between Ratings and Enrollments
Medicare Advantage Data
- Recall the Medicare Advantage repository, Medicare Advantage GitHub repository
Building the Data
Recall from the last two homework assignments (or snippets of those data, focusing on GA in 2022)
ga_plans_year– yearly GA plan panel (from contracts + enrollment).
ga_service_year– yearly GA service-area panel.
ga_landscape– GA plan characteristics (landscape, incl.premium_partc).
ga_penetration– GA penetration data.
- Now incorporate yearly
star-ratingsfiles
Understanding the Ratings Data
- Underlying measures change from year to year
- File
rating_variables.Rlists the relevant variables for each year partc_scorereflects specific star rating for Part C- Compilation of all underying measures plus additional adjustments
- For our purposes…just use the average of the underying measures
Contracts versus Plans
- Recall the difference between contractid and planid
- Star ratings are calculated at the contract level
- Applies to all plans operating under the same contractid
- Applies to all counties in which the same contractid is approved
- For our purposes…treat everything at the plan level
Example: Star Ratings in 2022
import pandas as pd
# Read in data
ga_ma_2022 = (
pd.read_csv("../data/output/ma-snippets/ga-ma-data-2022.csv")
.drop(columns=["partc_score"], errors="ignore")
)
ma_ratings = pd.read_csv("../data/output/ma-snippets/ga-ratings-2022.csv")
# Merge on contractid
ga_ma_full = ga_ma_2022.merge(ma_ratings, on="contractid", how="left")
# Variables used to construct the raw rating
rating_vars = [
"breastcancer_screen", "rectalcancer_screen", "flu_vaccine",
"physical_monitor", "specialneeds_manage", "older_medication", "older_pain",
"osteo_manage", "diabetes_eye", "diabetes_kidney", "diabetes_bloodsugar",
"ra_manage", "falling", "bladder", "medication", "statin", "nodelays",
"carequickly", "customer_service", "overallrating_care", "overallrating_plan",
"coordination", "complaints_plan", "leave_plan", "improve", "appeals_timely",
"appeals_review", "ttyt_available"
]
# Row-wise mean, ignoring missing values (na.rm = TRUE)
ga_ma_full["raw_rating"] = ga_ma_full[rating_vars].mean(axis=1, skipna=True)
# Keep the desired columns
ga_ma_full = ga_ma_full[
[
"contractid", "planid", "fips", "plan_type", "partd", "avg_enrollment",
"avg_eligibles", "avg_enrolled", "premium", "premium_partc",
"premium_partd", "rebate_partc", "ma_rate", "bid", "avg_ffscost",
"partc_score", "partcd_score", "raw_rating"
]
]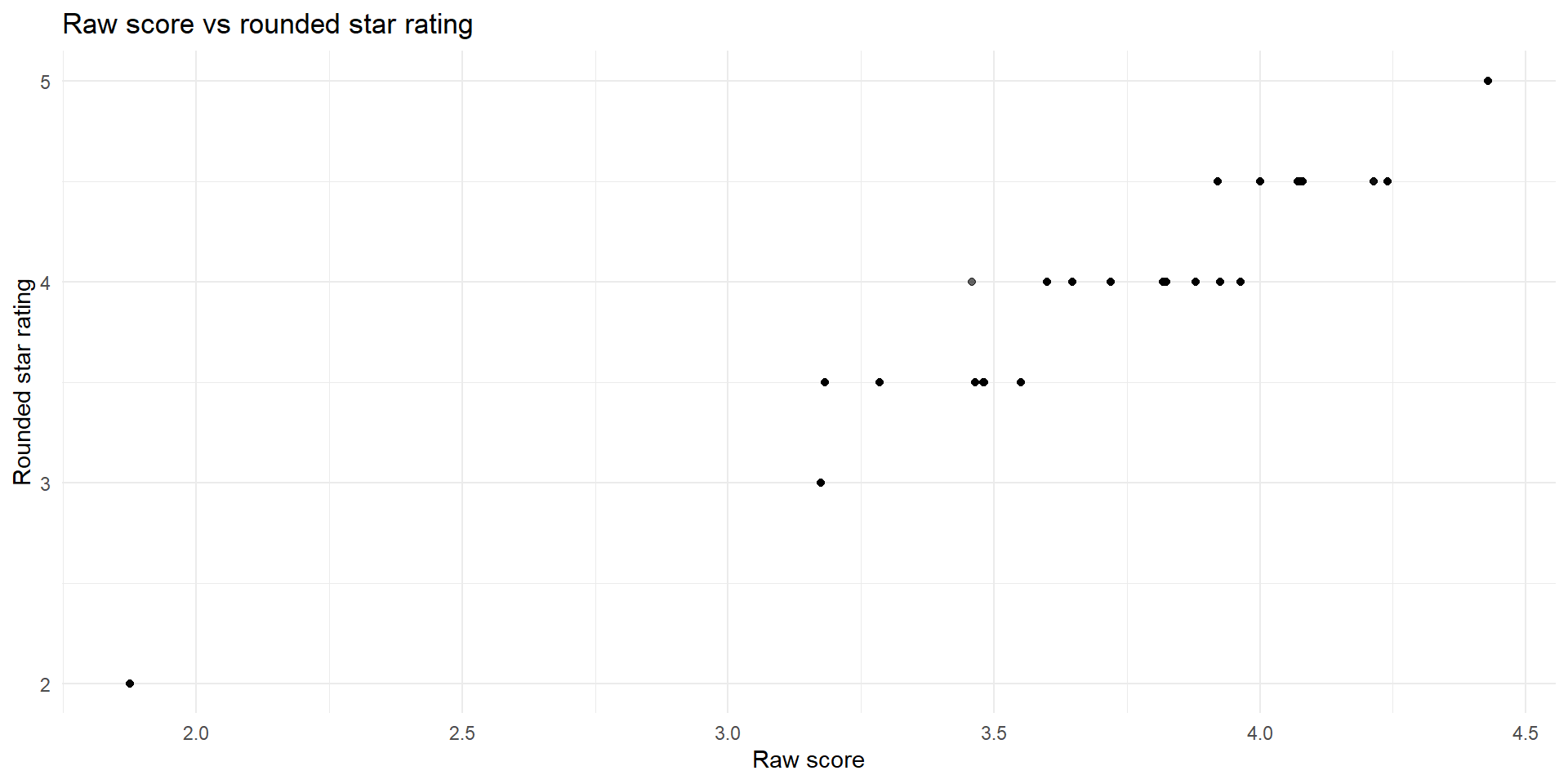
R Code
rounding_counts_3_75 <- ga_ma_full %>%
filter(
!is.na(raw_rating), !is.na(partc_score),
partc_score %in% c(3.5, 4.0),
raw_rating >= 3.5, raw_rating <= 4.0,
(raw_rating >= 3.75 & partc_score==4.0) | (raw_rating <= 3.75 & partc_score==3.5)
) %>%
mutate(
`Raw Score` = if_else(raw_rating >= 3.75, "≥ 3.75", "< 3.75"),
`Star` = if_else(partc_score == 4.0, "4.0", "3.5")
) %>%
count(`Raw Score`, `Star`, name = "Plans")
rounding_counts_3_75 %>%
kbl(
format = "html",
caption = "Ratings vs Raw Score"
) %>%
kable_styling(
bootstrap_options = c("striped", "hover", "condensed"),
full_width = FALSE,
position = "center"
)| Raw Score | Star | Plans |
|---|---|---|
| < 3.75 | 3.5 | 35 |
| ≥ 3.75 | 4.0 | 359 |
Enrollments and Star Ratings
Summary stats
Focus on enrollments and star ratings:
miss_n <- function(x) sum(is.na(x))
sum.vars <- ga_ma_full %>%
select("MA Enrollment" = avg_enrollment, "MA Eligibles" = avg_eligibles, "Star Rating" = partc_score,
"Raw Score" = raw_rating)
datasummary(All(sum.vars) ~ Mean + SD + miss_n + Histogram, data=sum.vars, fmt=2)| Mean | SD | miss_n | Histogram | |
|---|---|---|---|---|
| MA Enrollment | 153.62 | 456.49 | 0.00 | ▇ |
| MA Eligibles | 19141.94 | 28803.30 | 0.00 | ▇▂▁ |
| Star Rating | 3.97 | 0.52 | 3.00 | ▁▆▃▇ |
| Raw Score | 3.74 | 0.34 | 0.00 | ▂▄▁▇ |
import pandas as pd
import numpy as np
# 1. Subset and rename variables (analogous to select + renaming in R)
sum_vars = ga_ma_full.rename(columns={
"avg_enrollment": "MA Enrollment",
"avg_eligibles": "MA Eligibles",
"partc_score": "Star Rating",
"raw_rating": "Raw Score",
})[["MA Enrollment", "MA Eligibles", "Star Rating", "Raw Score"]]
# 2. Define missing-count function (miss_n)
def miss_n(x):
return x.isna().sum()
# 3. (Optional) simple text histogram for each variable
def hist_text(x, bins=10):
counts, _ = np.histogram(x.dropna(), bins=bins)
return " | ".join(map(str, counts))
# 4. Build summary table: Mean, SD, missing, and histogram
summary = pd.DataFrame({
"Mean": sum_vars.mean(),
"SD": sum_vars.std(),
"Missing N": sum_vars.apply(miss_n),
"Histogram": sum_vars.apply(hist_text), # drop this line if you don't want the histogram column
})
# 5. Optional: format to 2 decimal places for Mean and SD
summary[["Mean", "SD"]] = summary[["Mean", "SD"]].round(2)
summarySimple regression
Call:
lm(formula = avg_enrollment ~ factor(partc_score), data = ga_ma_full)
Residuals:
Min 1Q Median 3Q Max
-716.7 -134.4 -98.1 -30.6 9728.5
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 13.07 225.54 0.058 0.95378
factor(partc_score)3 40.11 227.50 0.176 0.86005
factor(partc_score)3.5 106.60 225.96 0.472 0.63713
factor(partc_score)4 187.91 226.26 0.831 0.40631
factor(partc_score)4.5 147.81 225.89 0.654 0.51294
factor(partc_score)5 715.46 237.12 3.017 0.00257 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 451.1 on 3275 degrees of freedom
(3 observations deleted due to missingness)
Multiple R-squared: 0.02584, Adjusted R-squared: 0.02436
F-statistic: 17.38 on 5 and 3275 DF, p-value: < 2.2e-16Potential endogeneity
- The star rating is a measure of quality
- Actual “quality” is not the same as “quality disclosure”
- Quality may be endogenous to enrollment (how?)
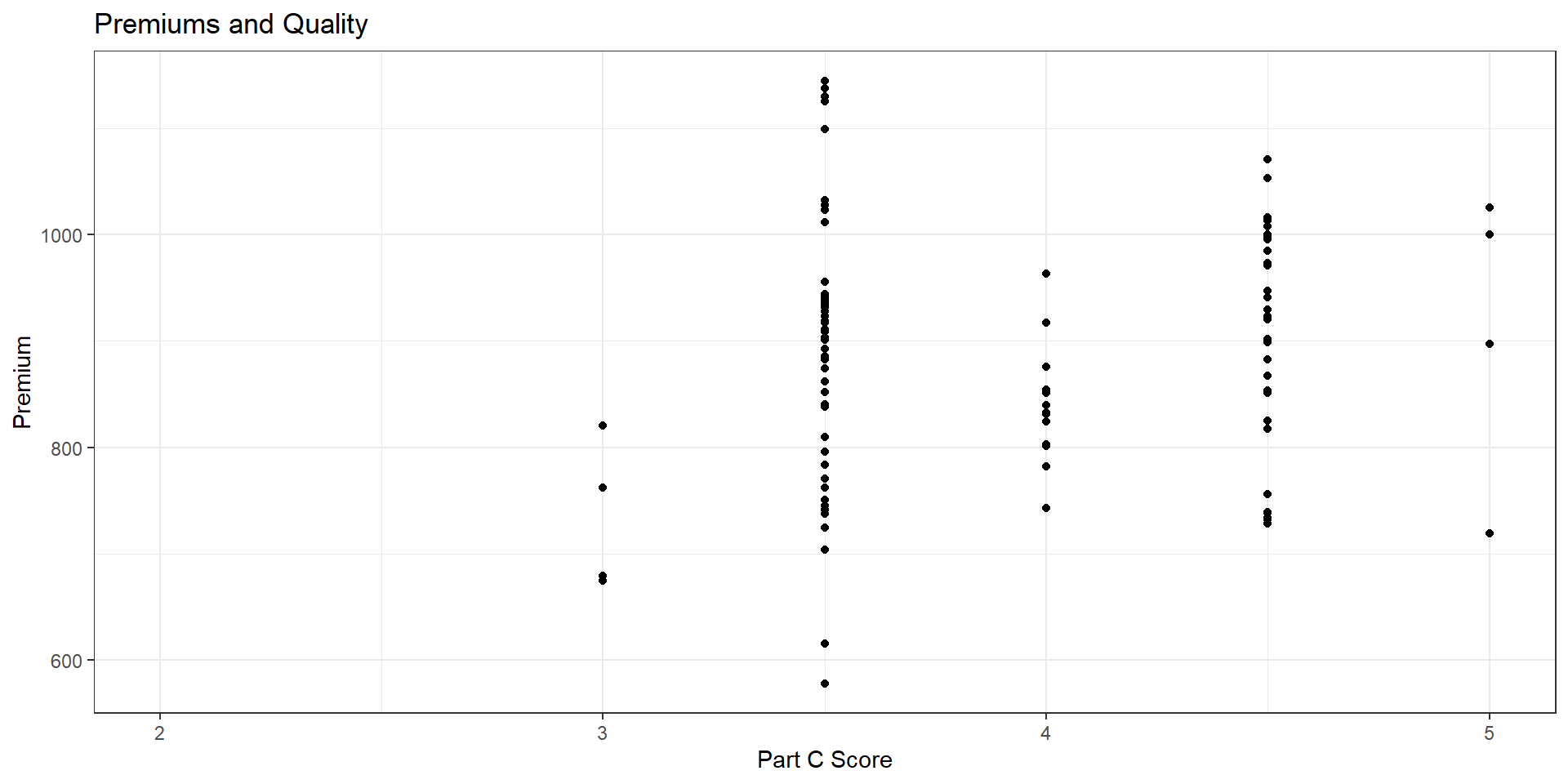
Call:
lm(formula = bid ~ factor(partc_score), data = ga_ma_full)
Residuals:
Min 1Q Median 3Q Max
-261.079 -71.049 9.059 70.648 305.049
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 750.240 6.714 111.739 < 2e-16 ***
factor(partc_score)3.5 89.295 7.392 12.079 < 2e-16 ***
factor(partc_score)4 80.448 7.853 10.244 < 2e-16 ***
factor(partc_score)4.5 181.633 7.284 24.936 < 2e-16 ***
factor(partc_score)5 119.729 17.831 6.715 2.21e-11 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 101.8 on 3272 degrees of freedom
(7 observations deleted due to missingness)
Multiple R-squared: 0.2319, Adjusted R-squared: 0.2309
F-statistic: 246.9 on 4 and 3272 DF, p-value: < 2.2e-16